Magnus Ducatus Lituaniae Project - BGA analysis project for the territories of former Grand Duchy of Lithuania. Admin: Vadim Verenich Co-admin: Leon Kull
My Blog List
Thursday, April 21, 2011
Experiments with relatedness
Yesterday i was investigating metodologies of pairwise IBD estimation in PLINK software. PLINK allows you to estimate genomewide IBD-sharing coefficients between seemingly unrelated individuals from whole-genome data. In a homogeneous sample, it is possible to calculate genome-wide IBD given IBS information, as long as a large number of SNPs are available (probably 1000 independent SNPs at a bare minimum; ideally 100K or more). The basic PLINK command for IBD calculations is
plink --file mydata --genome --min 0.05
which yields information useful for IBD estimation
FID1 Family ID for first individual
IID1 Individual ID for first individual
FID2 Family ID for second individual
IID2 Individual ID for second individual
RT Relationship type given PED file
EZ Expected IBD sharing given PED file
Z0 P(IBD=0)
Z1 P(IBD=1)
Z2 P(IBD=2)
PI_HAT P(IBD=2)+0.5*P(IBD=1) ( proportion IBD )
PHE Pairwise phenotypic code (1,0,-1 = AA, AU and UU pairs)
DST IBS distance (IBS2 + 0.5*IBS1) / ( N SNP pairs )
PPC IBS binomial test
RATIO Of HETHET : IBS 0 SNPs (expected value is 2)
Following the instructions from EMERGE Network article "Visualizing relatedness" and R graphic libraries (such as ggplot2) one can easily visualize Z1 and Z0, the proportion of markers identical by descent 1 and 0 respectively, for every pair of individuals in the dataset.
Example: Visualizing relatedness of project's "unrelated" sample of N=159
plink --file mydata --genome --min 0.05
which yields information useful for IBD estimation
FID1 Family ID for first individual
IID1 Individual ID for first individual
FID2 Family ID for second individual
IID2 Individual ID for second individual
RT Relationship type given PED file
EZ Expected IBD sharing given PED file
Z0 P(IBD=0)
Z1 P(IBD=1)
Z2 P(IBD=2)
PI_HAT P(IBD=2)+0.5*P(IBD=1) ( proportion IBD )
PHE Pairwise phenotypic code (1,0,-1 = AA, AU and UU pairs)
DST IBS distance (IBS2 + 0.5*IBS1) / ( N SNP pairs )
PPC IBS binomial test
RATIO Of HETHET : IBS 0 SNPs (expected value is 2)
Following the instructions from EMERGE Network article "Visualizing relatedness" and R graphic libraries (such as ggplot2) one can easily visualize Z1 and Z0, the proportion of markers identical by descent 1 and 0 respectively, for every pair of individuals in the dataset.
Example: Visualizing relatedness of project's "unrelated" sample of N=159
MDL historical portal
For those of you interested in MDL history, here is a link to the most informative MDL historical portal available at the moment. The portal is supported by Istorinės atminties akademija, Lietuvos Respublikos užsienio reikalų ministerija.
Wednesday, April 20, 2011
First results: Admixture unsupervised run
I've perfomed an unsupervised Admixture run on all of reference samples, shared by DW.
Monday, April 18, 2011
Leon Kull joins the team
It is my pleasure to announce that Mr.Leon Kull (of HIRsearch) has accepted the responsibilities of the project's co-admin.
An interim Report on The Progress of The MDL Project
Due to the effective cooperation on behalf of David Wesiolowski (of Eurogenes Project),the first batch of reference genotypes have already been merged to the project dataset.
I would like to express my deep appreciation for Zack (of Harappa DNA project) for his willingness to help me with some some data consversion issues.
I would like to express my deep appreciation for Zack (of Harappa DNA project) for his willingness to help me with some some data consversion issues.
Sunday, April 17, 2011
Introducing the Magnus Ducatus Lituaniae genetic project
The Magnus Ducatus Lituaniae [Grand Duchy of Lithuania] genetic project aims to analyze advanced genomic structure and levels of admixtures in individuals descending from the residents of the former Grand Duchy of Lithuania*
*[further denoted as MDL descendants].
- Project rationale
At the current moment, the project has one major goal: to build a sample (c.50-100 individuals) of MDL descendants (who have previously tested with 23andMe), which is statistically valid for inferring ancestry and admixture of individuals participated in project.
- The protection of privacy and confidentiality
Privacy - Your raw genetic data will not be shared with anyone and it will not be analyzed for anything other than ancestry or admixture.
Confidentiality - in order to protect confidentiality, a unique ID will be assigned to each participant of the project.
Under Directive 95/46/EEC, each participant will retain the right to get information on what data are recorded, how they are recorded and which methodologies are used.
- Criteria of participation
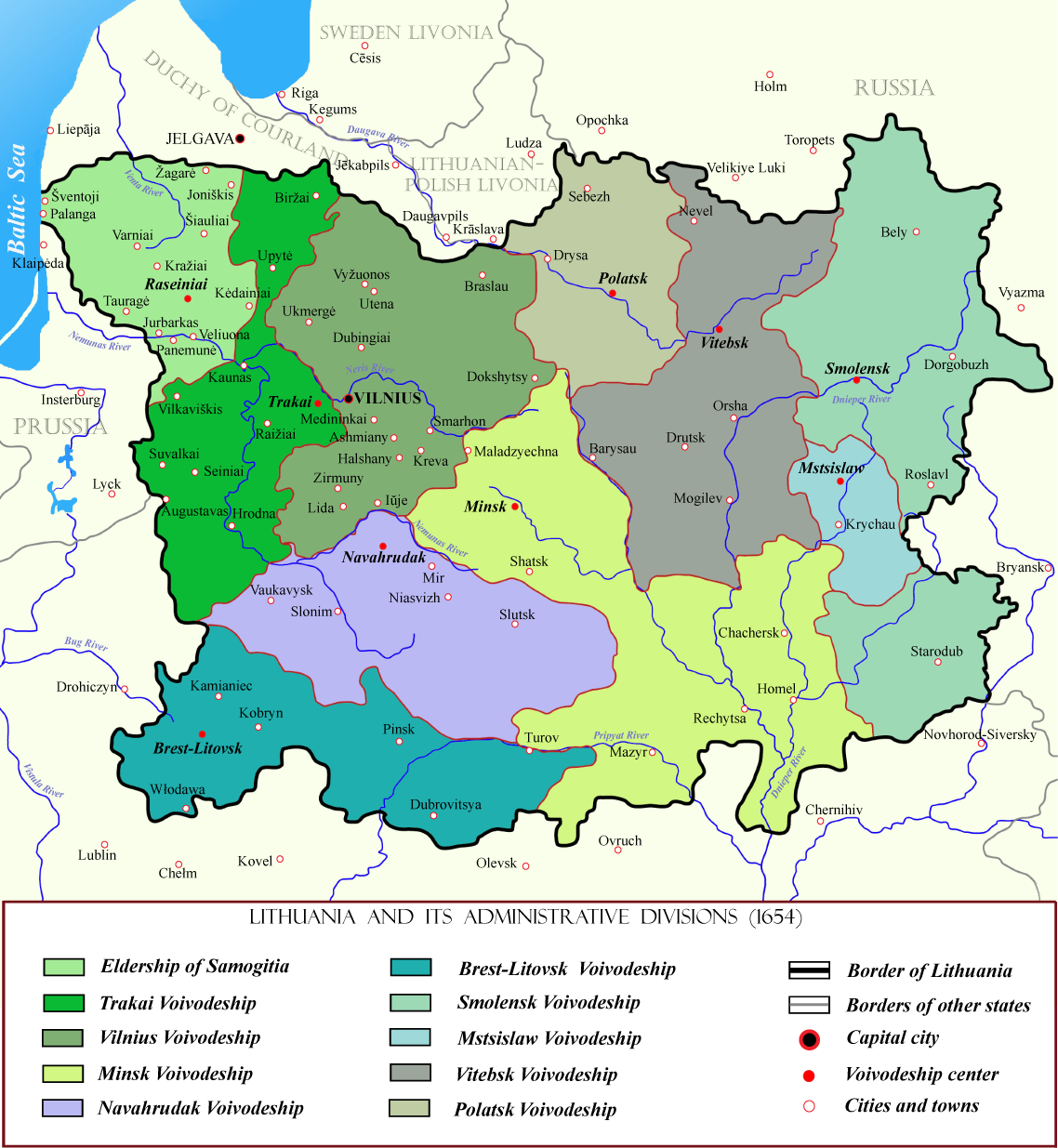
Generally,you are eligible to participate, if all of your 4th grandparents fall within the category of MDL descendants, i.e your grandparents came from the territory of the former Grand Duchy of Lithuania (1569-1795).
At present, i am looking for participants
- Belarusians
- Lithuanians (including Lithuanians from region of Samogitia and Prussia)
- Poles (from Lithuania propria and eastern parts of Poland - Mazovia and Podlasie)
- Latvians from Latgale (the former Inflanty Voivodeship -"polish" Livonia)
In order to participate, you should provide me with compressed genotype file from 23andMe.Send it to vadimverenich (a) gmail.com.
- Preliminary results of project
- You will receive a detailed Plink analysis, including the estimation of homozygous ROH (shared clusters and groups of homozygosity), possible Mendelian errors, extended LD-haplotypes (based on values of R2), shared IBD segments and IBS matrix (Plink format).
- You will receive your genotype file in haploid phase (this is a separate analysis demanding genotypes of your parents, so it will not be performed on a regular base) (Beagle or Merlin output format).
- You will receive the list of IBD segments detected by AISconvert (based on HIRsearch) and Germline software.
- Your data will be included in STRUCTURE analysis for detecting admixture clusters (see example of STRUCTURE output below)
- You will receive MDS plot showing your relative position to other participants of project (see example below)
- You will receive RHHmapper schemes showing the location of rare heterozygous and homozygous genotypes
Subscribe to:
Posts (Atom)